
Ajay Khanna
Bridging quantum mechanics and real-world impact through drug discovery, energy transfer, and sustainable technologies

Bridging quantum mechanics and real-world impact through drug discovery, energy transfer, and sustainable technologies
Computational chemistry at the intersection of molecular science and transformative applications
Virtual Screening & Design
FRET & Excitonic Coupling
Battery Materials
Sustainable Solutions
Calculating absorption and fluroscence spectra of molecules in explicit environment using ensemble Franck-Condon methods
A Python program designed automate MD trajectories containing dyes in solvents to hybrid QM/MM simulations.
An interactive Python GUI application allows you to visualize and manipulate molecules from an XYZ file.
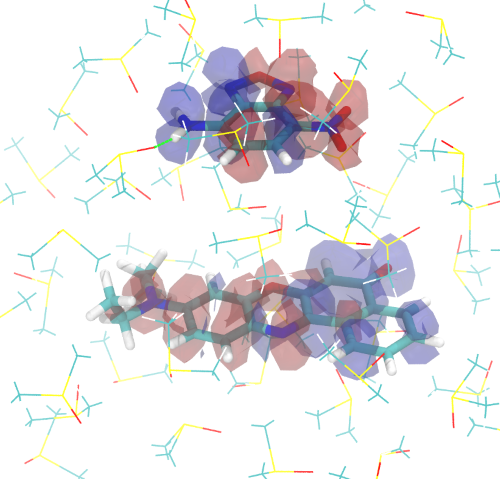
Computing Excitonic Coupling Between Molecules Using Various Excitonic Coupling Schemes

Convert Digital Object Identifiers (DOI) into BibTeX entries with a simple, user-friendly web app built using Streamlit!
Combining quantum mechanics, molecular simulations, and machine learning for real-world impact
Virtual screening, docking, and ADMET prediction for therapeutic design
Hybrid quantum/classical methods for excited states and optical properties
Periodic DFT for batteries, catalysts, and photovoltaic materials
ML/DL models for property prediction, QSAR, and molecular generation
High-performance computing and GPU acceleration for large-scale simulations
Large-scale compound screening, similarity search, and QSAR modeling
Research and development at the forefront of computational chemistry
Los Alamos National Laboratory (LANL)
Advisor: Dr. Sergei Tretiak
University of California, Merced
Computational Chemistry & Physics
Frontier Medicines
Computational Chemistry & Drug Discovery
Ajay Khanna, Jean-Huber Olivier, Sebastian Fernandez-Albertia, and Sergei Tretiak Peer-reviewed journal (2025 - Submitted) Under Review
|| Comprehensive study investigating structural disorder effects on electronic and optical properties in π-stacked perylene diimide aggregates, providing insights for organic photovoltaic design.
Ajay Khanna, Victor M. Freixas, Lei Xu, Jérémy R. Rouxel, Niranjan Govind, Marco Garavelli, Shaul Mukamel, and Sergei Tretiak J. Phys. Chem. Lett. 16 (2025) Journal Cover Top 15% Novelty
|| Delivered foundational insights into the structural origins of chiroptical signals, leading to a mechanistic understanding crucial for the rational design of functional molecular machines. Accepted without revision and selected for journal cover.
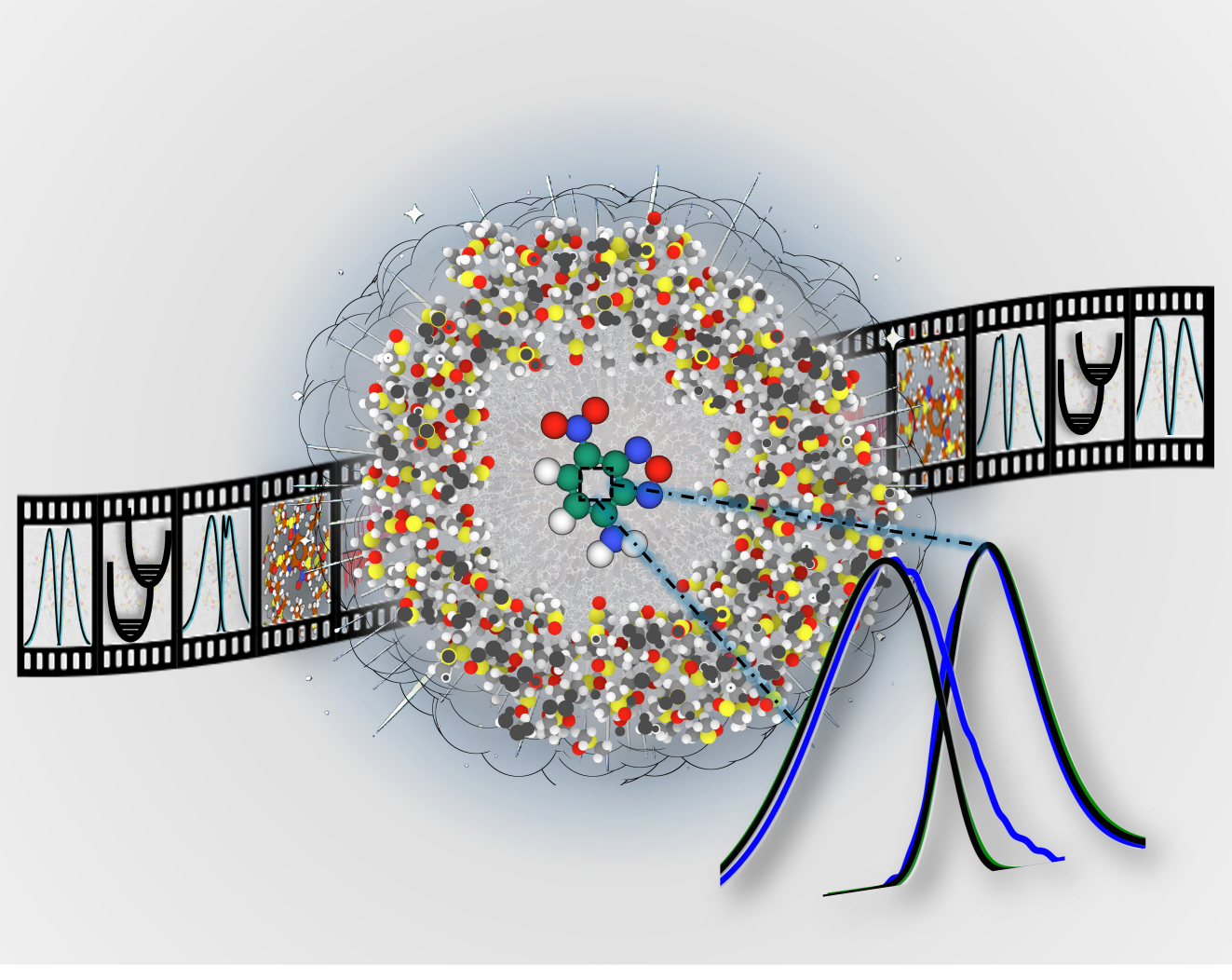
Ajay Khanna, Sapana V. Shedge, Tim J. Zuehlsdorff, Christine M. Isborn J. Chem. Phys. (2024)Citations: 6 Top 5-25% Novelty
|| Novel computational approach for accurate prediction of absorption and fluorescence spectra in solution, advancing spectroscopic analysis techniques.
Chiao-Yu Cheng, Nina Krainova, Alyssa Brigeman, Ajay Khanna, Sapana Shedge, Christine Isborn, Joel Yuen-Zhou, Noel C. Giebink Nature Comm. (2022) Citations: 22
|| Novel study on molecular polariton electroabsorption, opening new avenues in optoelectronics and quantum technology.
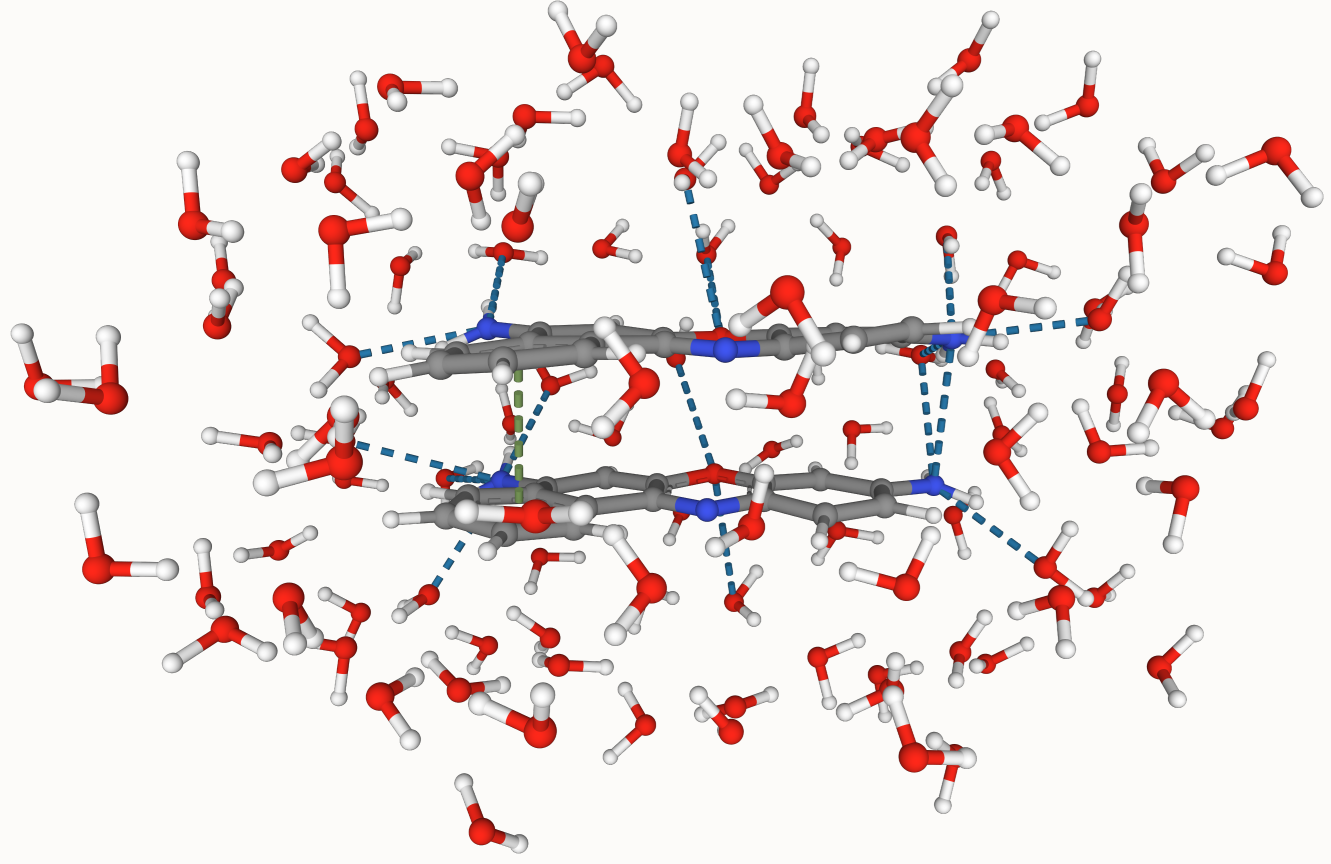
Sapana V. Shedge, Tim J. Zuehlsdorff, Ajay Khanna, Stacey Conley, Christine M. Isborn J. Chem. Phys. (2021) Citations: 15
|| Comprehensive exploration of environmental and vibronic effects in optical spectroscopy simulations, enhancing accuracy in molecular property predictions.
Christopher A. Myers, Shao-Yu Lu, Sapana Shedge, Arthur Pyuskulyan, Katherine Donahoe, Ajay Khanna, Liang Shi, and Christine M. Isborn The Journal of Physical Chemistry B 2024 Citations: 6
|| Systematic dissection of solvent effects on spectral line shapes, improving simulation accuracy by identifying the critical role of specific molecular interactions.
Rakesh Parida, G. Naaresh Reddy, Ajay Khanna, Gourisankar Roymahapatra, Santanab Giri Int. J. HIT. TRANSC: ECCN. Vol (2018)
|| Theoretical investigation of ligand-driven electron counting rules in germanium cluster complexes, contributing to understanding of main-group element chemistry.
Working with leading researchers and institutions worldwide to advance computational chemistry
Continuous learning and specialization in cutting-edge computational methods
Udemy
2024
Comprehensive training in computational methods for molecular representation, drug-target interactions, and structure-based drug design
Simplilearn
2022
Expertise in data analysis, visualization, and machine learning using Python ecosystem (NumPy, Pandas, Scikit-learn)
NVIDIA
2023
GPU-accelerated computing and parallel programming for high-performance scientific simulations

Getting started with running classical molecular dynamics simulations using Amber24

How to run ab initio molecular dynamics (AIMD) simulations in gas, implicit and explicit Enviroments

TeraChem for electronic structure calculations: Gas, implicit and explicit environments

Gaussian16 for electronic structure calculations: Gas, implicit and explicit environments

Click For More Tutorials on different types of MD and AIMD sims. Electronic structure, data analysis, quality images etc